WASHINGTON — For nearly 80 years, the VA has supported veterans with low-vision or blindness. Today, the VA’s Office of Blind Rehabilitation estimates that about 130,000 veterans are legally blind and another million have vision impairments that interfere with their daily lives.
Among older veterans, the leading causes of vision loss include age-related macular degeneration, glaucoma, cataracts, stroke, and diabetic retinopathy, while for those who served in Iraq and Afghanistan, vision issues often arise from blast-related brain injuries.
To meet the needs of all veterans with vision impairment, the VA established 13 Blind Rehabilitation Centers as well as 22 intermediate and advanced low-vision clinics and 23 advanced ambulatory low-vision clinics as well as the Visual Impairment Center to Optimize Remaining Sight (VICTORS) program. In addition, the VA and DoD jointly operate the Vision Center of Excellence and the Vision Impairment Services in Outpatient Rehabilitation (VISOR) program.
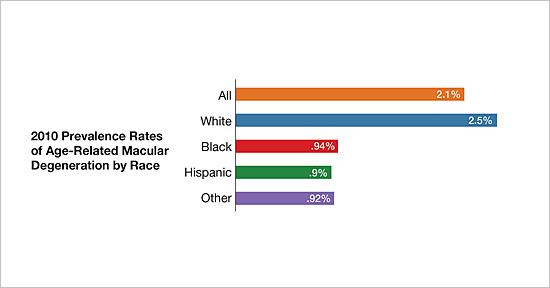
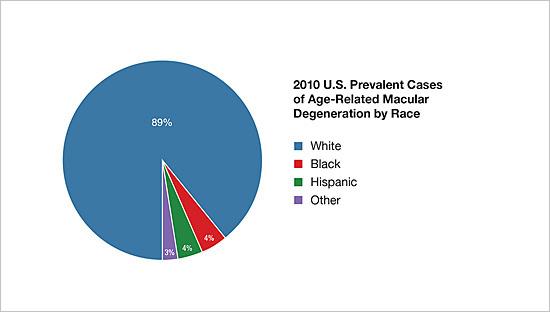
In addition to offering services for veterans with visual impairments, the VA has long been a leader in research into treatments for common eye diseases, particularly age-related macular degeneration (AMD), the leading cause of vision impairment in older veterans. “An analysis of ICD codes relative to macular degeneration approximates that 158,000 veterans were diagnosed with a form of macular degeneration in fiscal year 2021 at Veterans Affairs facilities, from a little over 1.53 million veterans receiving eye care at VA facilities,” VA Spokesperson Gina Jackson told U.S. Medicine. “Thus, nearly 10% of the veterans who were seen in VA eye care clinics during fiscal year 2021 were diagnosed with macular degeneration.”
The most significant risk factors for AMD are age, northern European ancestry, and genetic, while smoking is the primary modifiable risk factor.
Age-related Macular Degeneration
AMD causes loss of central vision, characterized by blurring of details in images as a result of damage to the macula from a reduced supply of oxygen and nutrients as blood vessels in the retina harden. AMD interferes with the ability to clearly see faces, read, drive, adjust to low lighting, see colors, and maintain many hobbies. It may affect one or both eyes.
About 10% of individuals aged 66 to 74 have macular degeneration, rising to 30% in those aged 75 to 85 years. In addition to age, risk factors include certain genetic mutations, family history of AMD, elevated blood pressure, light iris color, obesity, smoking, and high cholesterol. Several infectious agents have also been implicated in the development of AMD including Chlamydia pneumoniae, cytomegalo virus, and human herpesvirus-6A.
There are two types of AMD, dry and wet. In dry AMD, small yellow deposits called drusen appear between the layers of the retina. As more and larger drusen develop, the light-sensitive cells in the macula become thinner and eventually die. Dry AMD progresses slowly and has no symptoms in its early and intermediate stages. Patients with dry AMD have a 4% to 12% chance per year of progressing to wet AMD. About 30% of individuals with AMD in one eye will develop AMD in the other eye within four years.
In wet or neovascular macular degeneration, abnormal new blood vessels start to grow under the retina. These fragile blood vessels easily hemorrhage, leading to swelling and scarring of the macula. Wet AMD is always considered late stage and can progress very quickly.
Digging Into the Genetics
Because age and genetic mutations are the most significant risk factors for AMD, researchers at the VA and elsewhere have sought to understand which mutations confer the greatest risk and why. John B. Harley, MD, PhD, of the Cincinnati VAMC and his colleagues at other leading research institutions recently published a study that used elastic data-driven genetic encoding (EDGE) to detect the significant genetic interactions in simulated genetic models of the AMD phenotype, among others. They then performed a multi-encoding genome-wide association study (GWAS) for each phenotype to identify significant interactions of single nucleotide polymorphisms (SNPs).1
The method found genome-wide significant results for AMD with six SNPs using all four traditional encoding methods, which included four SNPs in the complement factor H (CFH) gene. A SNP in the age-related maculopathy susceptibility 2 (ARMS2) gene was also detected.
Previous studies have found that mutations in the CFH and ARMS2 genes account for more than half of AMD’s heritability. Additionally, “while activation of the complement system is central to controlling microbial infection, it also is a well-recognized player in AMD pathogenesis,” said Australian researchers in Front Cell Neuroscience. “This is demonstrated by the identification of complement signaling protein constituents in drusen— a pathological hallmark of AMD.”2
The EDGE interaction analysis plus other encoding methods found two significant SNP-SNP interactions. “The top SNP-SNP model for this phenotype was rs2336502 (alpha: 0.39; MAF:0.33; pseudogene LOC100996886)–rs6695321 (alpha: 0.52; MAF: 0.36; intron, CFH),” the authors noted. “Another significant interaction model also included rs2336502, which was identified as interacting with rs5993 (alpha: 0.28; MAF: 0.26; intron, coagulation factor XIII B chain (F13B)).”
“The results of the simulation study and epistasis analysis in data from the eMERGE Network and UK Biobank indicate that there are examples of SNPs with a nonadditive genetic model,” Harley and colleagues wrote, “which demonstrates the need for encoding methods beyond additive, including EDGE.”
- Hall MA, Wallace J, Lucas AM, Bradford Y, Verma SS, Muller–Myhsok B, et al. Novel EDGE encoding method enhances ability to identify genetic interactions. PLoS Genet. 2021;17(6): e1009534. https://doi.org/10.1371/journal.pgen.1009534
- Too LK, Hunt N, Simunovic MP. The Role of Inflammation and Infection in Age-Related Neurodegenerative Diseases: Lessons From Bacterial Meningitis Applied to Alzheimer Disease and Age-Related Macular Degeneration. Front Cell Neurosci. 2021 Mar 26;15:635486. doi: 10.3389/fncel.2021.635486. PMID: 33867940; PMCID: PMC8044768.