
Early in the COVID-19 pandemic, staff Sgt. DeAngela Cranor, assigned to the Javits New York Medical Station, monitors a COVID-19 patient in the facility’s intensive care unit. Now, research using veterans has identified genetic links between SARS-C0V-2 infection severity and certain medical conditions that are known risk factors for severe virus cases. U.S. Navy photo by Chief Mass Communication Specialist Barry Riley.
PHILADELPHIA — From the earliest days of the pandemic, it was evident that people with certain medical conditions faced an increased risk of severe COVID-19. As the pandemic progressed, research began to identify certain variants in specific human genes that are associated with more severe infection with SARS-C0V-2.
Now, a study led by VA researchers has identified genetic links between COVID-19 severity and certain medical conditions that are known risk factors for severe virus cases.
Reasoning that variants may be associated with other medical conditions that might already be understood, the researchers analyzed data from the VA Million Veteran Program (MVP), one of the largest and most racial and ethnically diverse cohorts in the United States, with genetic information linked with electronic health records (EHR) on more than 650,000 veterans.
“We selected generic variants associated with severe COVID-19 and conducted a phenome-wide association study, a type of analysis that let you examine the association between variants and a wide range of clinical conditions reported in the [electronic health records],” said Anurag Verma, PhD, of the Division of Translational Medicine and Human Genetics at the University of Pennsylvania, one of the study’s authors.
Their analysis found variants associated with risk factors for severe COVID-19 manifestations such as Type 2 diabetes, ischemic heart disease and coagulopathies, Verma explained. Among respiratory conditions, idiopathic pulmonary fibrosis and chronic alveolar lung disease shared genetic risk factors with severe COVID-19.1
“One of the results that stood out to us was that a variant near the TYK2 gene associated with severe COVID-19 was also associated with a reduced risk of autoimmune conditions, such as psoriasis and lupus,” Verma said. MVP’s racial ethnical diversity gave the researchers a unique opportunity to investigate associations within each genetic ancestry, he noted. “A variant in the LMNA gene was associated with neutropenia—a condition with abnormally low counts of a type of white blood cell—in African ancestry but not in European. It is particularly interesting, as LMNA gene is linked with a broad spectrum of cardiomyopathies such as dilated cardiomyopathies and familial atrial fibrillation. However, the association with neutropenia has not been previously reported.”
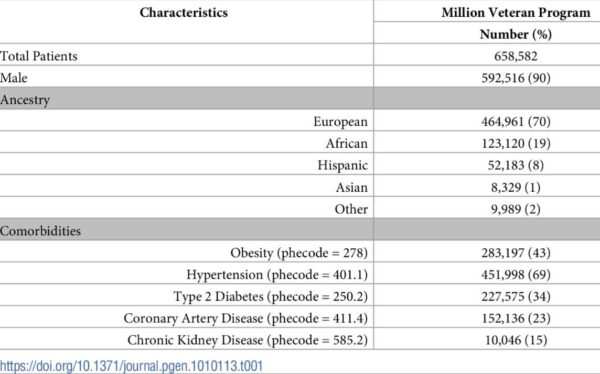
Click To Enlarge: Patient characteristics of Million Veteran Program participants. Source: PLOS Genetics 18(4): e1010113.
Rheumatologist and lead author Katherine Liao, MD, MA, of the VA Boston Healthcare System said she was initially surprised at the high number of immune-mediated conditions that came up from the PheWAS. “When we tested the association between genes related to severe COVID-19 and screened to see if they were also associated with about 1,800 other conditions, many were autoimmune conditions such as psoriasis or systemic lupus erythematosus,” she noted.
“The nature of the relationships was also initially puzzling,” Liao pointed out. “For example, gene A was associated with developing severe COVID-19, but gene A was also associated with a reduced risk of developing psoriasis, an immune-mediated skin disease. Some of these genes are linked to how our immune system functions.”
She noted that the immune system is constantly balancing fighting infection but not getting too active and attacking itself, which is what happens in autoimmune conditions. “Thus, in hindsight, it may not be surprising that individuals who manifest with more severe COVID—some of whom may have weaker immune systems—would also be less likely to attack selves and have a lower risk of developing some autoimmune conditions.”
Liao said she considers the finding as the first step to “get a lay of the land” and provide hypothesis-generating data. “This study provided the foundation to understand which conditions share genes with a predilection to develop severe COVID,” she said. “Starting with these conditions where we often have decades of knowledge and known effective therapies, we can then use this information to target follow-up studies aimed at improving the management and treatment of COVID across a diverse population.”
The authors concluded that the “PheWAS of genetic variants reported to associate with severe COVID-19 demonstrated shared genetic architecture between COVID-19 severity and known underlying risk factors for both severe COVID-19 and poor COVID-19 outcomes, rather than susceptibility to other viral infections. Overall, the associations observed were generally consistent across genetic ancestries, with the exception of a stronger association with neutropenia among veterans of [African] and [Hispanic] ancestry and not [European]. Notably, only few respiratory conditions had a shared genetic association with severe COVID-19.”
They added that, among these, “variants associated with a reduced risk for severe COVID-19 had an opposite association, with reduced risk for inflammatory and fibrotic pulmonary conditions. Similarly, other divergent associations were observed between severe COVID-19 and autoimmune inflammatory conditions, shedding light on the concept of the fine balance between immune tolerance and immunodeficiency. This balance will be important when considering therapeutic targets for COVID-19 therapies, where pathways may control both inflammation and the viral host response.”
Liao stressed the study would not have been possible without the VA and the data from veterans. “I can’t underscore enough that one of the successes of MVP is the willingness of our veterans to continue to serve through participating in this study,” Liao said. “Studies using genetics typically require large populations to be able to see associations between genes and disease. The combined data of 650K veterans across the country placed VA and MVP in a position to have a large enough dataset to relatively quickly study which conditions share genes related to severe manifestations of COVID-19.”
- Verma A, Tsao N, Thomann L, Ho YL, et. al. A Phenome-Wide Association Study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program. medRxiv [Preprint]. 2021 Oct 15:2021.05.18.21257396. doi: 10.1101/2021.05.18.21257396. Update in: PLoS Genet. 2022 Apr 28;18(4):e1010113. PMID: 34642702; PMCID: PMC8509103.


